Primer design module#
The circtools primex module is a highly specialized primer design tool tailored specifically for circRNA experiments.
circtools primex is able to design primer pairs in batches of hundreds of circRNAs based on circRNAs detected with circtools detect, but can also work on lists with specific circRNA isoforms or even entirely without any preliminary data purely based on the FASTA sequence of the circRNA.
The circtools primex module is based on the equally named R package
Required tools and packages#
circtools primex depends on R, several R packages, and BioPython:
R packages:
primex
formattable
kableExtra
dplyr
RColorBrewer
colortools
Python libraries:
BioPython>=1.71
All R package as well as Python dependencies are installed during the circtools installation.
General usage#
A call to circtools primex --help shows all available command line flags:
usage: circtools [-h] -d DCC_FILE -g GTF_FILE -f FASTA_FILE [-O {mm,hs}]
[-s SEQUENCE_FILE] [-o OUTPUT_DIR] [-T EXPERIMENT_TITLE]
[-t GLOBAL_TEMP_DIR] [-G GENE_LIST [GENE_LIST ...]]
[-p PRODUCT_SIZE [PRODUCT_SIZE ...]]
[-i ID_LIST [ID_LIST ...]] [-j {r,n,f}] [-b]
circular RNA primer design
optional arguments:
-h, --help show this help message and exit
Input:
-d DCC_FILE, --dcc-file DCC_FILE
CircCoordinates file from DCC / detect module
-g GTF_FILE, --gtf-file GTF_FILE
GTF file of genome annotation e.g. ENSEMBL
-f FASTA_FILE, --fasta FASTA_FILE
FASTA file with genome sequence (must match
annotation)
-O {mm,hs}, --organism {mm,hs}
Organism of the study (used for primer BLASTing), mm =
Mus musculus, hs = Homo sapiens
-s SEQUENCE_FILE, --sequence SEQUENCE_FILE
FASTA file containing the circRNA sequence (exons and
introns)
Output options:
-o OUTPUT_DIR, --output OUTPUT_DIR
Output directory (must exist)
-T EXPERIMENT_TITLE, --title EXPERIMENT_TITLE
Title of the experiment for HTML output and file name
Additional options:
-t GLOBAL_TEMP_DIR, --temp GLOBAL_TEMP_DIR
Temporary directory (must exist)
-G GENE_LIST [GENE_LIST ...], --genes GENE_LIST [GENE_LIST ...]
Space-separated list of host gene names. Primers for
CircRNAs of those genes will be designed.E.g. -G
"CAMSAP1" "RYR2"
-p PRODUCT_SIZE [PRODUCT_SIZE ...], --product-size PRODUCT_SIZE [PRODUCT_SIZE ...]
Space-separated range for the desired PCR product.
E.g. -p 80 160 [default]
-i ID_LIST [ID_LIST ...], --id-list ID_LIST [ID_LIST ...]
Space-separated list of circRNA IDs. E.g. -i
"CAMSAP1_9_135850137_135850461_-"
"CAMSAP1_9_135881633_135883078_-"
-j {r,n,f}, --junction {r,n,f}
Should the forward [f] or reverse [r] primer be
located on the BSJ? [Default: n]
-b, --no-blast Should primers be BLASTED? Even if selected yes here,
not more than 50 primers willbe sent to BLAST in any
case.
Designing primers with circtools primex#
A sample call to primex using the Jakobi et al. 2016 data generated with circtools detect requires as only external parameter the Fasta sequence of the reference genome in order to obtain DNA sequences for the primer design process.
# obtain reference genome (if not already downloaded)
wget ftp://ftp.ensembl.org/pub/release-90/fasta/mus_musculus/dna/Mus_musculus.GRCm38.dna.primary_assembly.fa.gz
# obtain annotation (if not already downloaded)
wget ftp://ftp.ensembl.org/pub/release-90/gtf/mus_musculus/Mus_musculus.GRCm38.90.gtf.gz
# unzip
gzip -d Mus_musculus.GRCm38.dna.primary_assembly.fa.gz
gzip -d Mus_musculus.GRCm38.90.gtf.gz
# run circtools primex, design primer for gene Ryr2
circtools primex -d DCC/CircCoordinates -f Mus_musculus.GRCm38.dna.primary_assembly.fa -g Mus_musculus.GRCm38.90.gtf -O mm -G Ryr2 -T "Ryr2 primer"
Start parsing GTF file
Start merging GTF file
extracting flanking exons for circRNA # 1 Ryr2_13_11680966_11688013_-
extracting flanking exons for circRNA # 2 Ryr2_13_11690292_11700868_-
extracting flanking exons for circRNA # 3 Ryr2_13_11718370_11730486_-
extracting flanking exons for circRNA # 4 Ryr2_13_11737722_11745759_-
extracting flanking exons for circRNA # 5 Ryr2_13_11749436_11785141_-
extracting flanking exons for circRNA # 6 Ryr2_13_11759671_11772579_-
extracting flanking exons for circRNA # 7 Ryr2_13_11759671_11779268_-
extracting flanking exons for circRNA # 8 Ryr2_13_11759671_11785141_-
extracting flanking exons for circRNA # 9 Ryr2_13_11769852_11785141_-
extracting flanking exons for circRNA # 10 Ryr2_13_11779185_11801925_-
extracting flanking exons for circRNA # 11 Ryr2_13_11824274_11853190_-
extracting flanking exons for circRNA # 12 Ryr2_13_11868117_11885538_-
Sending 92 primers to BLAST
This may take a few minutes, please be patient.
Writing results to /tmp/Ryr2_primer.html
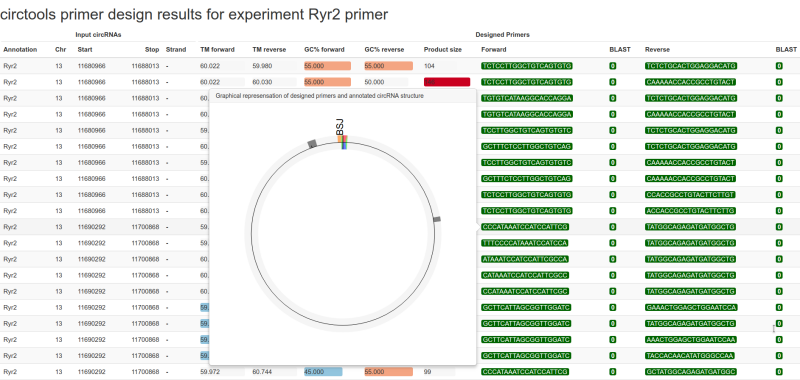
circtools primex takes a few seconds to process the input data and sends the generated primers pairs to the web-based BLAST service of the NCBI in order to give the user hints about potential unwanted off-site targets. The output is written to a HTML file which can be opened with any browser.
Sample of the HTML output generated by circtools primex#